|
|
| |
 |
Single Molecule AFM Force
Spectroscopy - PHYSICS OF MOLECULAR RECOGNITION
Contact: Dr. Volker Walhorn |
| |
|
|
| |
|
Research Topic: The physical mechanisms
of specific, non-covalent intermolecular binding is quantitatively
investigated by single-molecule (dynamic) force spectroscopy with
atomic force microscopy (AFM), optical tweezers (OT) and magnetic tweezers (MT). Here, specific molecular forces,
elasticities, kinetic
reaction rate constants (lifetimes) and the energy landscape
of the molecular binding potential is measured by AFM at the single-molecule
level in vitro on isolated but functional complexes. Nowadays, these specific
interactions can be scrutinized in a quantitative manner at
the sensitivity level on single-point mutations
(nucleic acids, amino acids) allowing single-molecule affinity
ranking in a broad affinity range of 0,1 mM-1 fM revealing
distinct differences in the binding properties and mechanisms. Over the last years, special emphasis has been put on
so-called molecular catch-bond systems that were observed when investigating the interplay of human sulfatates (Sulf1, Sulf2) with
glycosaminoglycans (heparan sulfate, heparin, dermatan sulfate, ...) in cell signaling cascades. In addition, we recently explored the homophilic interaction mechanisms between
desmosomal desmoglein-2 (DSG2) and mutations thereof that play an important role for the integrity of mechanically active cells like
cardiomyocytes. |
| |
|
|
| |
|
 |
| |
|
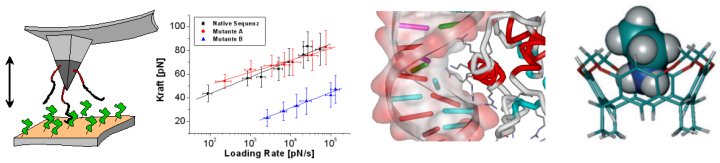
Images: Single molecule force spectroscopy
methodology (left), dynamic AFM force-spectroscopy-plots (middle left), atomistic model for peptide-DNA interaction (middle right), and the result of a quantum chemistry calculation for a supramolecular
host-guest-complex (right). |
| |
|
|
| |
|
References: ... see section Publications |
Last updated: 06.03.2017
|
|



