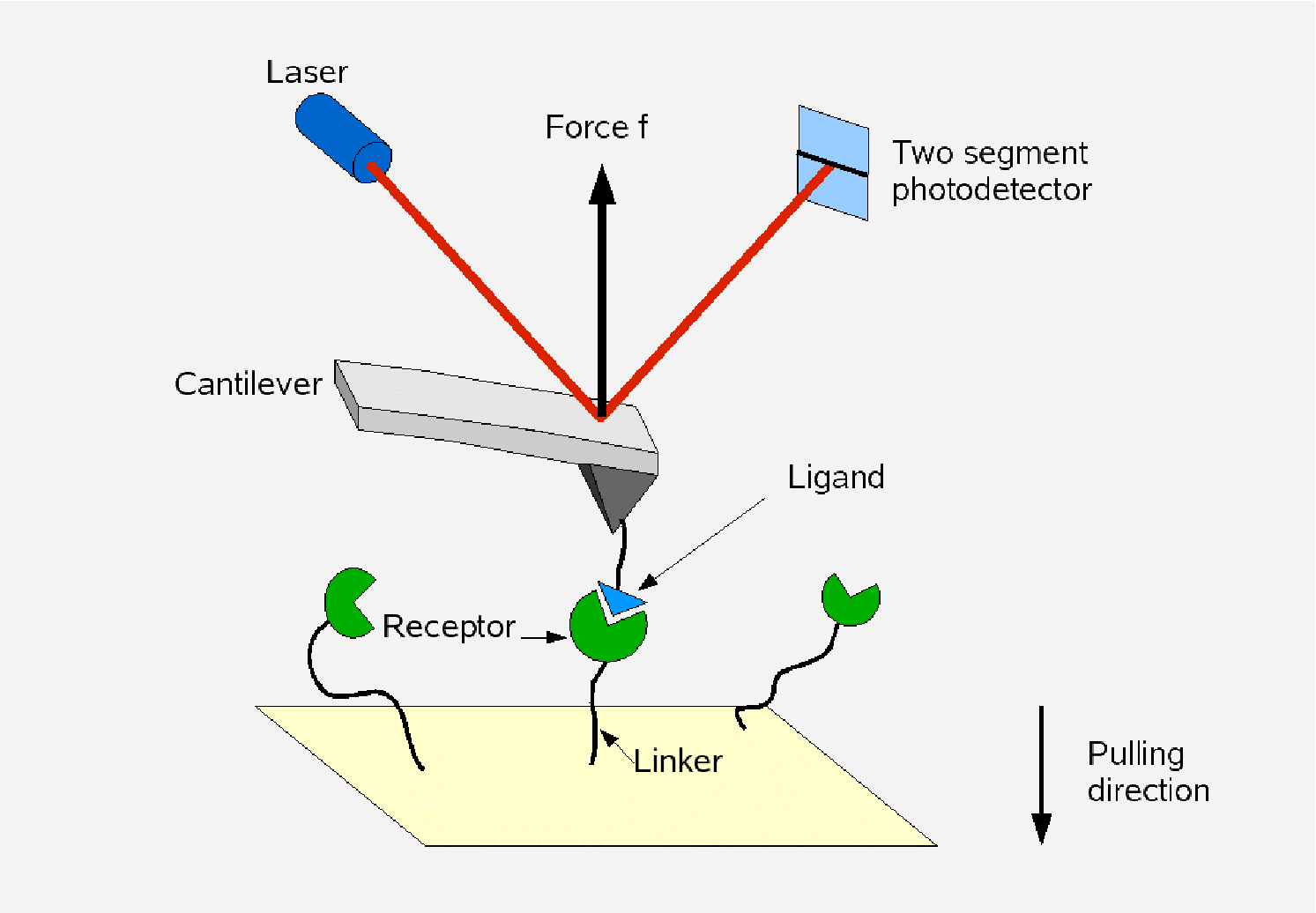
| Participating group members: | Mykhaylo Evstigneev, Sebastian Getfert, Peter Reimann |
| Main cooperation partners: | Dario Anselmetti
(Biophysics and applied Nanoscience
group at the University of Bielefeld;
Robert Ros, Alexander Fuhrmann (Arizona State University) |

M. Evstigneev and P. Reimann
Dynamic force spectroscopy: Optimized data analysis
Phys. Rev. E 68, 045103(R) (2003)
M. Raible, M. Evstigneev, P. Reimann, F. W. Bartels, and R. Ros
Theoretical analysis of dynamic force spectroscopy experiments on ligand-receptor complexes
J. Biotech. 112, 13 (2004)
M. Raible and P. Reimann
Single-molecule force spectroscopy: Heterogeneity of chemical bonds
Europhys. Lett. 73, 628 (2006)
M. Raible, M. Evstigneev, F. W. Bartels, R. Eckel, M. Nguyen-Duong,
R. Merkel, R. Ros, D. Anselmetti, and P. Reimann
Theoretical Analysis of Single-Molecule Force Spectroscopy Experiments: Heterogeneity of Chemical Bonds
Biophys. J. 90, 3851 (2006)
S. Getfert and P. Reimann
Optimal evaluation of single-molecule force spectroscopy experiments
Phys. Rev. E 76, 052901 (2007)
A. Fuhrmann, D. Anselmetti, R. Ros, S. Getfert, and P. Reimann
Refined procedure of evaluating experimental single-molecule force spectroscopy data
Phys. Rev. E 77, 031912 (2008)
S. Getfert, M. Evstigneev, and P. Reimann
Single-molecule force spectroscopy: Practical limitations beyond Bell's model
Physica A 388, 1120 (2009)
S. Getfert and P. Reimann
Hidden Multiple Bond Effects in Dynamic Force Spectroscopy
Biophys. J. 102, 1184 (2012)
Related projects:
Friction phenomena on the nanometer scale
Decay of complex metastable states
Last modified on 2012-03-14